Solving the C. difficile’s armour - while developing my own

About me
In November 2017, I landed in the UK from Brazil. Doing a PhD abroad had been an aspiration since I decided to pursue a career in Science, back in 2009 when I started my undergraduate degree in Biomolecular and Physical Sciences, at University of São Paulo (Brazil). As much as I wanted to go abroad for the full doctorate, finding the right project wasn’t an easy process - and getting a scholarship to fund it complicated things even further. My interest in Structural Biology flourished during my undergraduate internships, and since then I understood that this was my field of interest. Applying my knowledge to Microbiology, especially targeting antimicrobial resistance, stemmed from a big question - why and how can microorganisms escape human natural defences, and how do they fool antibiotics?
Practical aspects of the project
Even though I had some previous experience solving the crystal structure of lysozyme, real world crystal problems can involve crystallising the uncrystallisable, dealing with pathologic crystals, only managing to collect diffraction data at low resolution and finally getting to the big boss: solving the model.
Crystallisation of SlpA had previously been optimised by my supervisor, Dr. Paula Salgado, over the course of several years. However, due to its inherent ability to form 2D lattices, SlpA would readily crystallise into thin plates, leading to a highly anisotropic diffraction pattern with very weak signal.
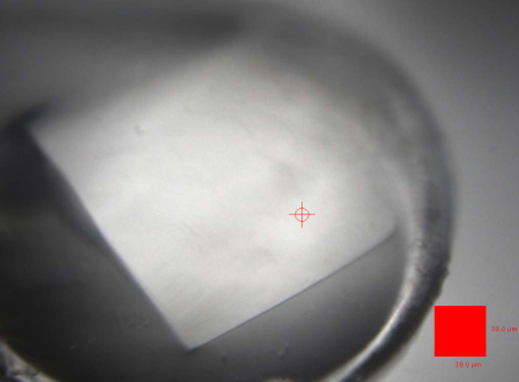
My job was to try and optimise SlpA crystallisation and data collection further, but, ultimately, determine the structure using highly pathologic and low resolution datasets. Although S-layers are quite a common feature in bacteria, the proteins that assemble them aren’t conserved at all. This means that there wasn’t any model available that would make phasing in molecular replacement an easy job - and experimental phasing with Se-Met was proven to be a rather dangerous experiment (as Paula and Rob may one day be able to report!). Phasing the full length SlpA crystal model was only possible by expressing, crystallising and solving the substructures of SlpA - the interacting domains LID/HID and the SLPL/HID. This work was carried out by my brilliant colleague Dr. Anna Barwinska-Sendra, who managed to take care of “baby PhD student” Paola while conducting her own research. The key datasets used to solve the structures now published were collected on the same day: the 28th of September 2018. This trip involved the three of us (Paula, Anna and me) travelling to Diamond Light Source for an overnight shift, testing and collecting data from hundreds of SlpA crystals. Out of the hundreds of crystals, only a handful of them diffracted nicely, and finally only two were good enough to allow for a crystal model solution.
Roadblocks
Fast forwarded two years of manual model building and refinements, going back and forth with the structure and learning a lot about low-medium resolution, in early 2020 the models of SlpA from 2 strains, a mutant and the substructures were ready for deposition and manuscript was on its way. Just as we were closing the story, Covid-19 also decided to close down the university - and ultimately, the world. For me, lockdowns in 2020 were not too harsh: Spring/Summer 2020 saw some very nice weather in the UK and moving to a “work from home” routine allowed me to work while enjoying the flowers I planted on my windowsill. I know I sound like a dreamer, but at that point, everything was fine - even when we thought the world was about to end.
Happy ending
Despite the odds, I handed in my PhD thesis in September 2021. Scientific writing (in your second language) is a really challenging job. Of course, I had careful guidance from my supervisor and a couple of other colleagues reviewing my writing. Keeping some sanity during the write-up process was a fine balance between trying to keep a routine (and yes, we were in lock down again!), trying to exercise and trying not to overeat. And yes, apparently, I survived my PhD! And the result of all the hard work is finally shared with the world in Nature Communications.
But I’ll tell you my secret: I couldn’t survive without a few heroes that helped me get through.
