Nature Communications, 2022
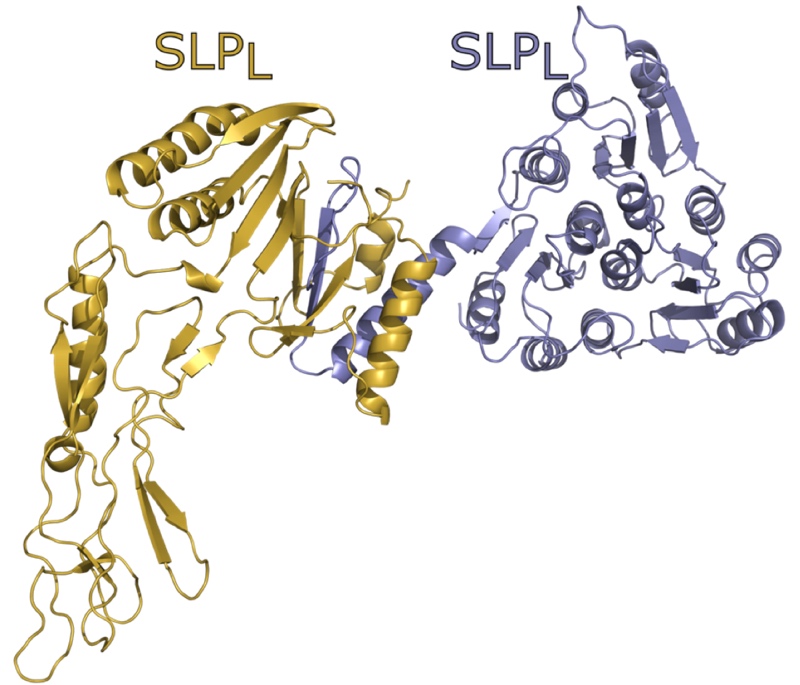
Structure and assembly of the S-layer in C. difficile
Abstract Many bacteria and archaea possess a two-dimensional protein array, or S-layer, that covers …
Read More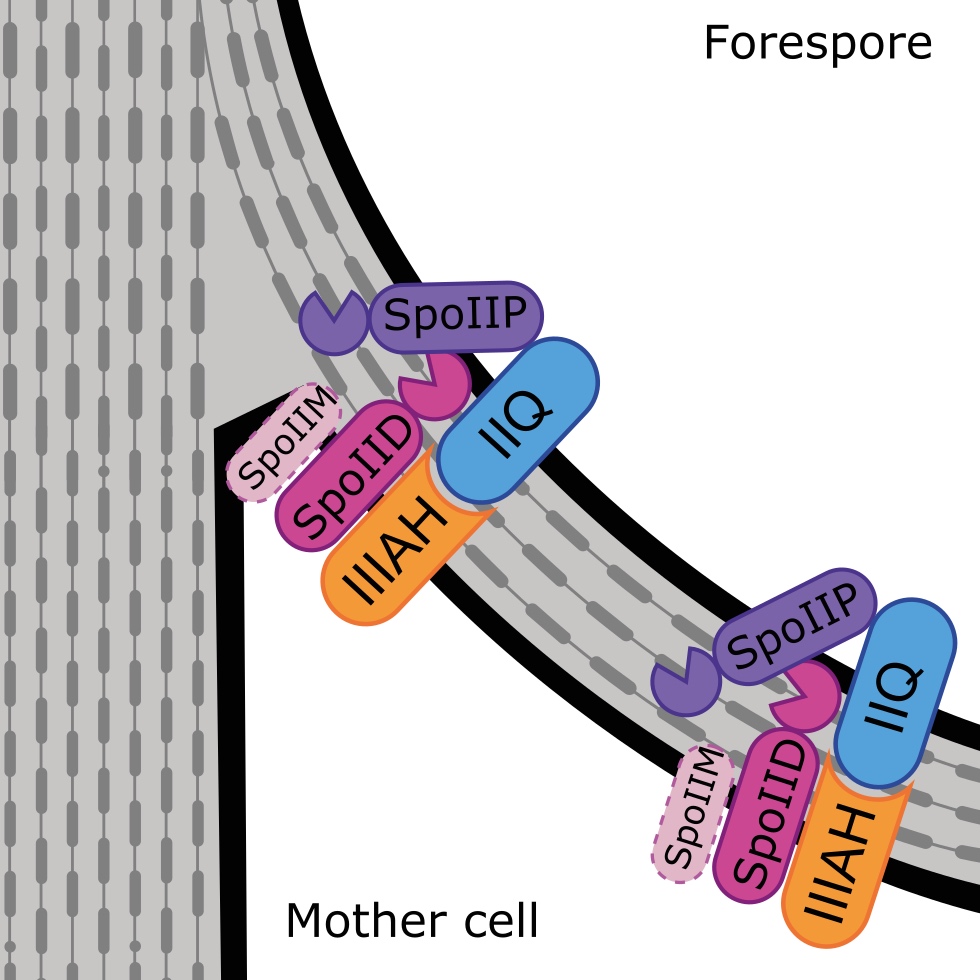
Molecular Microbiology, 2018
Peptidoglycan degradation machinery in Clostridium difficile forespore engulfment
Abstract Clostridium difficile remains the leading cause of antibiotic-associated diarrhoea in …
Read More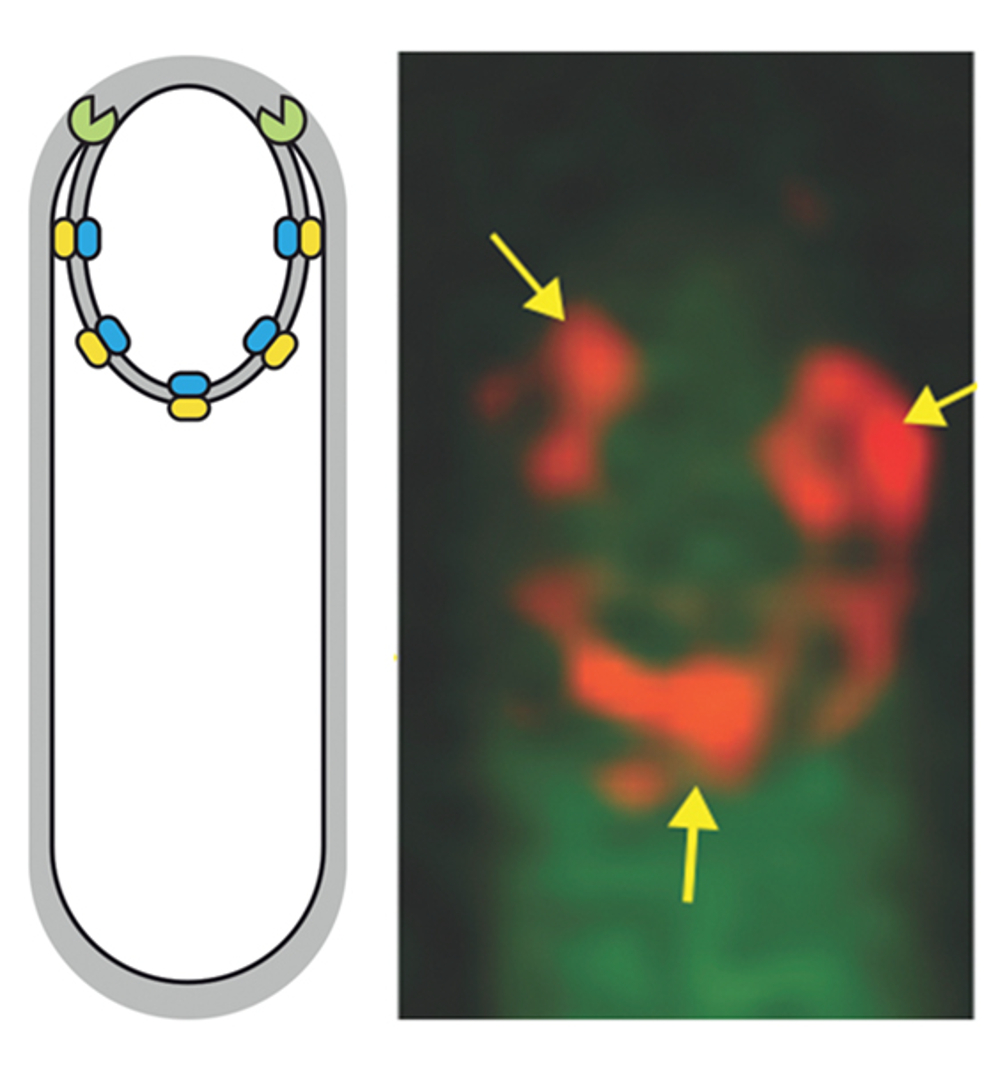
Molecular Microbiology, 2016
The SpollQ-SpolllAH complex of Clostridium difficile controls forespore engulfment and late stages of gene expression and spore morphogenesis
Abstract Engulfment of the forespore by the mother cell is a universal feature of endosporulation. …
Read More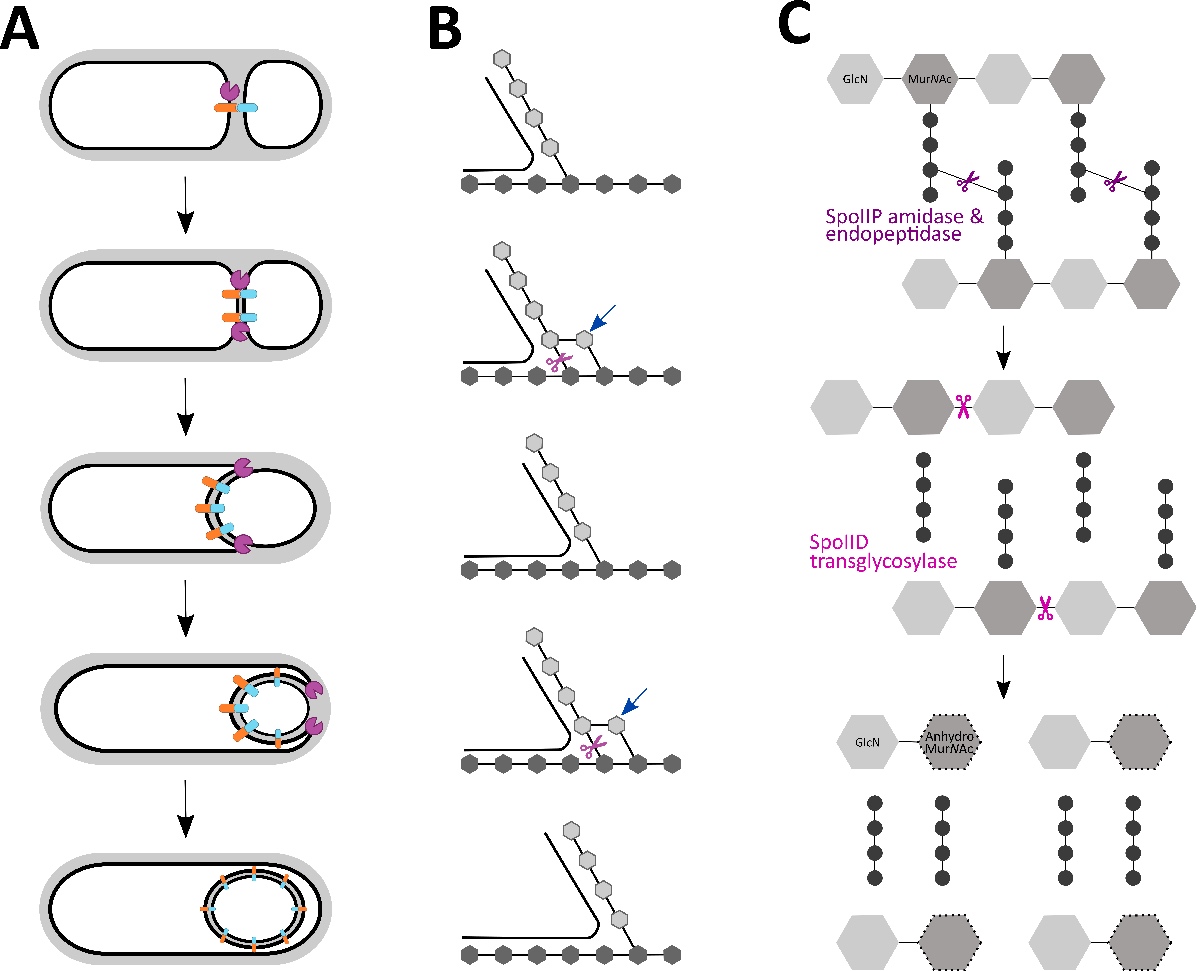
Anaerobe, 2019
The engulfasome in C. difficile: Variations on protein machineries
Abstract Clostridioides difficile infection (CDI) continues to be a substantial healthcare burden, …
Read More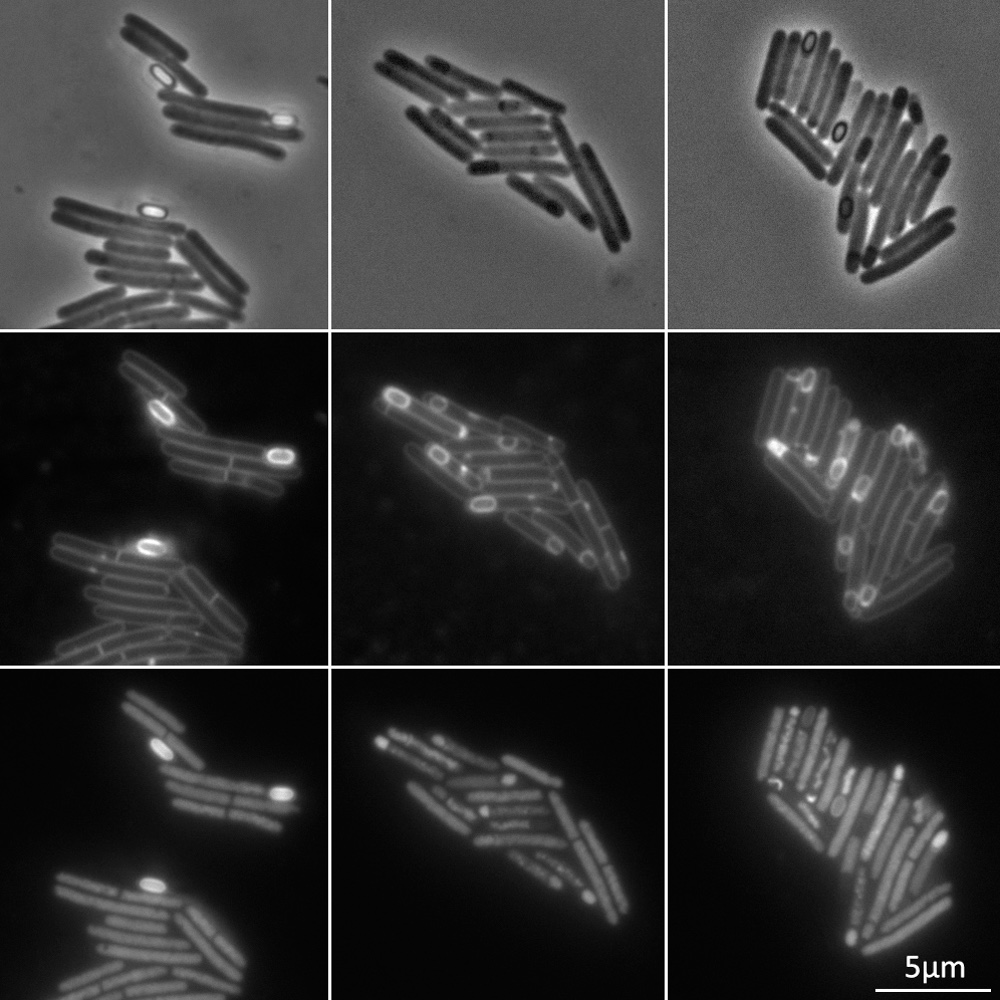
Frontiers in Microbiology, 2017
Inducible expression of spo0A as a universal tool for studying sporulation in Clostridium difficile
Abstract Clostridium difficile remains a leading nosocomial pathogen, putting considerable strain on …
Read More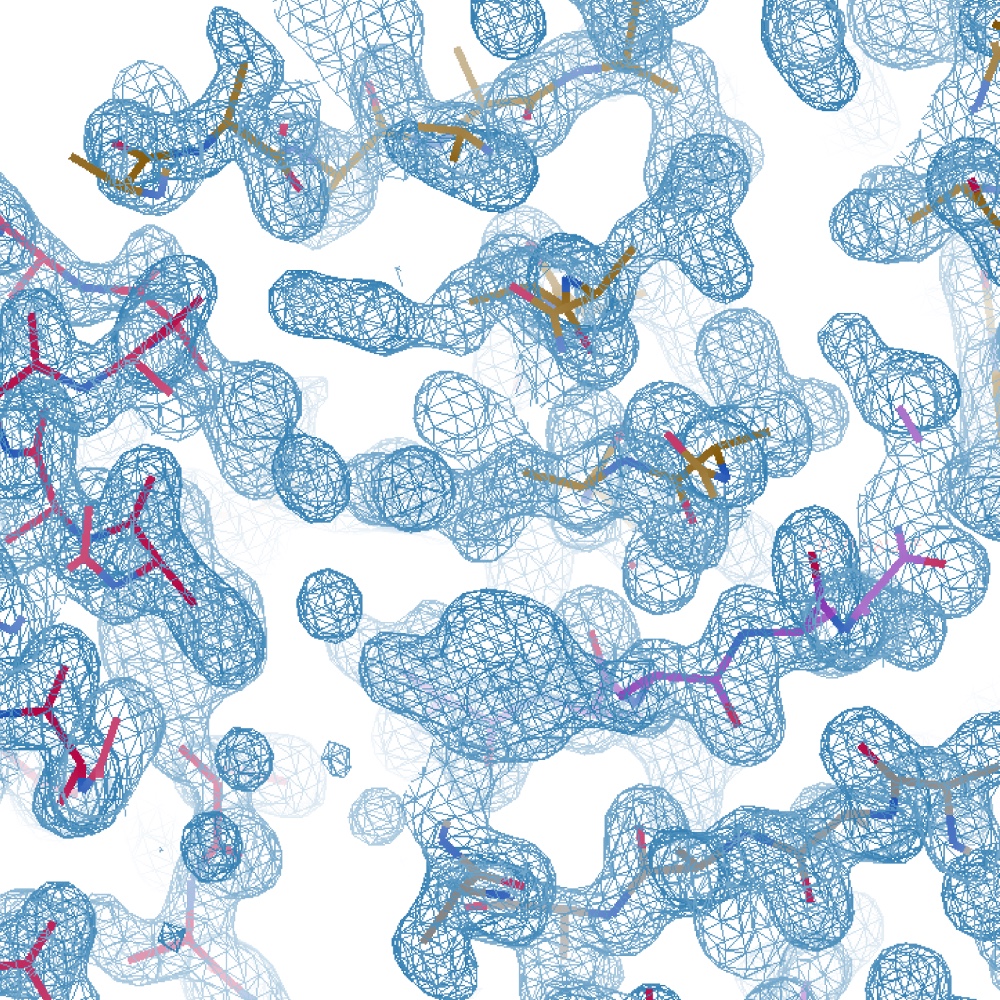
Acta Cryst D76, 2020
A practical overview of molecular replacement: Clostridioides difficile PilA1, a difficult case study
Abstract Many biologists are now routinely seeking to determine the three-dimensional structures of …
Read MoreOther publications
- Julius C., Salgado P.S., Yuzenkova Y. (2020) Metabolic cofactors NADH and FAD act as non-canonical initiating substrates for a primase and affect replication primer processing in vitro. Nucleic Acids Research. 48(13):7298-7306.
- Herrero-de-Dios, C., Day, A.M., Tillmann, A.T., Stravroula, L.K., Stead, D., Salgado, P.S., Quinn, J., Brown, A.J.P., (2018) Redox regulation, rather than stress-induced phosphorylation, of a Hog1 mitogen-activated protein kinase modulates its nitrosative-stress-specific outputs, mBio, 9 2e02229-1.
- Warren, A.J., Crawshaw, A.D., Trincao, J., Aller, P., Alcock, S., Nistea, I., Salgado, P.S., Evans, G. (2015) In vacuo X-ray data collection from graphene-wrapped protein crystals. Acta Cryst D71: 2079-88
- Lin, J., Oh, S.H., Jones, R., Garnett, J.A., Salgado, P.S., Rusnakova, S., Matthews, S.J., Hoyer, L., Cota, E. (2014) The peptide-binding cavity is essential for Als3-mediated adhesion of Candida albicans to human cells. J Biol Chem M114. 547877
- Douse, C.H., Green, J.L., Salgado, P.S., Simpson, P.J., Thomas, J. C., Langsley, G., Holder, A.A., Tate, E.W., Cota, E. (2012) Regulation of the Plasmodium motor complex: phosphorylation of Myosin A Tail Interacting Protein (MTIP) loosens its grip on MyoA. J Biol Chem 287:36968-77
- Liu, B., Garnett, J.A., Lee, W.C., Lin, J., Salgado, P.S., Taylor, J., Xu, Y., Lambert, S., Cota, E., Matthews, S. (2012) Promoting crystallisation of the Salmonella enteritidis fimbriae 14 pilin SefD using deuterium oxide. Biochem Biophys Res Commun 421, 208-13.
- Salgado, P.S.*, Yang, R.*, Taylor, J.D., Buchnell, L., Jones, R., Hoyer, L., Matthews, S.J., Simpson, P., Cota, E. (2011) Structural basis for the broad specificity to host-cell ligands by the pathogenic fungus Candida albicans. PNAS 108:15775-9 F1000 highlight
- Taylor, J.D., Zhou, Y., Salgado, P.S., Patwardhan, A., McGuffie, M., Pape, T., Grabe, G., Ashman, E., Constable, S.C., Simpson, P.J., Lee, W., Cota, E., Chapman, M.R., Matthews, S.J. (2011) Atomic Resolution Insights into Curli Fibre Biogenesis. Structure 19:1307-16
- Salgado, P.S.*, Taylor, J.D.*, Cota, E. And Mathews, S.J. (2011) Extending the phasing power of diselenide bonds: SeCys SAD phasing of CsgC using a non-auxotrophic strain. Acta Cryst D67, 8–13.
- Salgado, P.S., Yang, R., Rowan, F., Cota, E. (2011) Expression, crystallization and preliminary X-ray data analysis of NT-ALS9, a fungal adhesin from Candida albicans. Acta Cryst F67(Pt 4):467-70.
- Poranen, M.M.*, Salgado, P.S.*, Laurila, M.R.L., Wright, S., Bamford, D.H., Stuart, D.I., Grimes, J.M. (2008) Structural explanation for the role of Mn2+ in the activity of Φ6 RNA-dependent RNA polymerase. Nucleic Acid Research 36:6633-44.
- Salgado, P.S., Laurila, M.R.L., Makeyev, E.V., Bamford, D.H., Stuart, D.I., Grimes, J.M. (2006) The structure of an RNAi polymerase links RNA silencing and transcription. PloS Biol 4 :e434.
- Salgado P.S., Walsh, M.A., Laurila, M.R.L., Stuart, D.I. and Grimes, J.M. (2005) Going soft and SAD with manganese. Acta Cryst D61:108-11.
- Laurila, M.R.L.*, Salgado, P.S.*, Makeyev, E.V., Nettelship, J., Stuart, D.I., Grimes, J.M., Bamford, D.H. (2005) Gene silencing pathway RNA-dependent RNA polymerase of Neurospora crassa: yeast expression and crystallization of selenomethionated QDE-1 protein. J Struct Biol 149(1):111-5.
- Laurila, M.R.L.*, Salgado, P.S.*, Stuart, D.I., Grimes, J.M., Bamford, D.H. (2005) Back-priming mode of Phi6 RNA-dependent RNA polymerase. J Gen Virol 86:521 – 526.
- Salgado, P.S., Makeyev, E.V., Butcher, S.J., Bamford, D.H., Stuart, D.I., Grimes, J.M. (2004) The structural basis for RNA specificity and Ca2+ inhibition of an RNA-dependent RNA polymerase. Structure 12:307-16.
Book chapters
- Makeyev, E.V.*, Salgado, P.S.* (2005) Structure, function and evolution of virus and cell-encoded RNA-dependent RNA polymerases. In Recent Advances in RNA virus replication (K. Hefferon, Ed.), 2006, ISBN 81-7895-214-9, Research Signpost/Transworld Research Network (invited contribution)