S-layer

Many bacteria and archaea possess a two-dimensional protein array, the S-layer, that covers the cell surface and plays crucial roles in cell physiology. In C. difficile, the S-layer has been implicated in colonisation and inflammation.
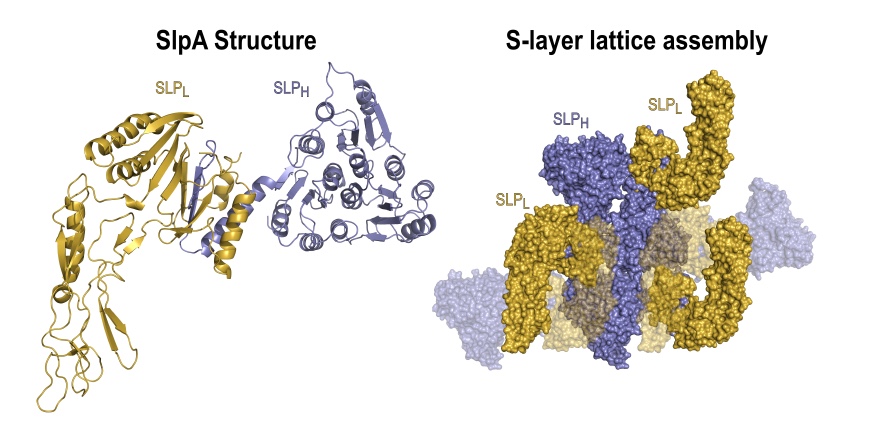
We have determined the structure and assembly of SlpA, the main S-layer component in C. difficile, which is post-translationally cleaved into two S-layer proteins (SLPs): the high molecular weight, SLPH, and low molecular weight, SLPL. These subunits then form a complex (H/L) that is incorporated in the S-layer.
Key findings in our Nature Communications paper (2022):
- SLPH forms a trimeric prism arrangement
- SLPL extends towards the environment
- The interacting domains form an intricate paper-clip motif that links the two SLPs
- Tiling of the SLPH creates the 2D array
- Interactions of SLPL cover the gaps between SLPH
- SlpA forms a very tightly packed lattice, replicated in crystals and in situ S-layer reconstructions
- Electron diffraction of empty S-layer ghosts provides a map of the complete S-layer in the cells
- The crystal structure of H/L complex mimics the organisation of the S-layer in the cells, providing a model for S-layer assembly in C. difficile.
We aim to continue to use a combination of structural, biochemical, biophysical and computational methods to further understand the role and dynamics of the S-layer.
Image by Lizah van der Aart